Abstract
Background: TP53 mutations in acute myeloid leukemia (AML) are associated with complex karyotype, high incidence of minimal residual disease (MRD), and high risk of relapse (Döhner et al., 2017; Giacomelli et al., 2018). While numerous novel treatment regimens, including the combination of the BCL2 inhibitor venetoclax (VEN) and hypomethylating agents (HMA), have emerged as partially effective treatments and resulted in higher remission rates in patients with TP53-mutant AML, full clearance of the mutant TP53 clone is rarely achieved and the majority of patients relapse (Short et al., 2021; Takahashi et al., 2016). The mechanisms responsible for response and relapse in TP53-mutant AML remain unclear and investigating novel mechanisms is critical to develop more effective therapies.
Results: In order to shed light on the defective p53 signaling pathways underlying TP53 mutant AML, and to better understand mechanisms of resistance, we performed RNA-sequencing (RNA-seq) on FACS-sorted subpopulations using samples collected from TP53-mutant or TP53-wt high-risk AML patients. Samples were collected at diagnosis (DX) and post-treatment (POSTTX) (total number of samples n= 67, TP53-mutant=36, TP53-wt=31). Diagnostic samples include bulk AML, leukemic stem cells (LSCs), and post-treatment samples including bulk mononuclear cells (MNCS) and patient specific MRD (total n= 67, DX_Bulk=15, DX_LSCs=15, POSTTX_MNCs=14, POSTTX_MRD=23). Differential gene expression analysis of TP53-mutant samples indicates a positive enrichment of the following pathways: G2/M checkpoint, MYC targets, and mitotic spindle, among others. We focused here on genes associated with TP53-mutant AML enriched pathways, and identified a key regulator of centriole biogenesis, one of E2F targets: Polo-like kinase 4 (PLK4) as a potential target highly expressed in TP53-mutant AML samples . Previous publications showed that PLK4 is transcriptionally repressed by p53 and induces apoptosis upon RNAi silencing (Fischer et al., 2014; Li et al., 2005). Here we show that TP53-mutant AML samples lack the p53-dependent PLK4 repression and have higher levels of PLK4 compared to TP53-wt AML.
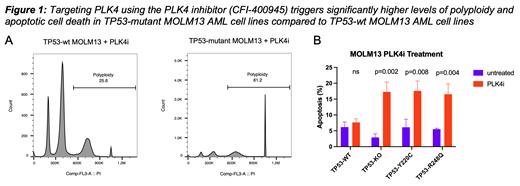
To test the rigor of this finding, we interrogated the Munich Leukemia Laboratory (MLL) data base and analyzed their clinically annotated (e.g. karyotype, survival, complete blood counts, previous treatments ... etc) RNA-seq dataset of 726 AML samples (TP53-mutant=72, TP53-wt=654). TP53-mutant AML samples consistently showed significant PLK4 upregulation (p= 0.0003). We analyzed PLK4 expression and its correlation with TP53 mutations in The Cancer Dependency Map project dataset (1375 cell lines in 35 different types of cancers) (p= 0.004). Furthermore, we found significantly higher PLK4 protein levels in TP53-mutant AML MOLM13 cell lines when compared with syngeneic TP53-wt AML MOLM13 cells. Experimentally, we found that PLK4 inhibition using 25nM CFI-400945 in TP53-mutant AML MOLM13 cell lines triggers polyploidy > 2-fold higher than in TP53-wt AML MOLM13 cell lines 72 hours post treatment (Fig.1A p< 0.0001). Finally, we show that polyploidy is not reversible after drug removal and results in significantly increased levels of apoptotic cell death in TP53-mutant AML MOLM13 cells (Fig.1B).
Conclusion: Our data suggest that TP53-mutant AML expresses higher levels of PLK4 in comparison to TP53-wt AML, and targeting PLK4 triggers polyploidy and apoptotic cell death in TP53-mutant AML. A clinical trial is ongoing testing the efficacy of PLK4 inhibition (CFI-400945) in AML (Clinical Trial ID: NCT04730258, TWT-202).
Issa: Kura Oncology: Consultancy, Research Funding; Syndax Pharmaceuticals: Research Funding; Novartis: Consultancy, Research Funding. Borthakur: Takeda: Membership on an entity's Board of Directors or advisory committees; ArgenX: Membership on an entity's Board of Directors or advisory committees; Ryvu: Research Funding; Astex: Research Funding; University of Texas MD Anderson Cancer Center: Current Employment; Protagonist: Consultancy; Novartis: Consultancy, Membership on an entity's Board of Directors or advisory committees; GSK: Consultancy. Konopleva: Ascentage: Other: grant support, Research Funding; Novartis: Other: research funding pending, Patents & Royalties: intellectual property rights; Stemline Therapeutics: Research Funding; KisoJi: Research Funding; Eli Lilly: Patents & Royalties: intellectual property rights, Research Funding; Sanofi: Other: grant support, Research Funding; AstraZeneca: Other: grant support, Research Funding; Ablynx: Other: grant support, Research Funding; AbbVie: Consultancy, Honoraria, Other: Grant Support, Research Funding; F. Hoffmann-La Roche: Consultancy, Honoraria, Other: grant support; Reata Pharmaceuticals: Current holder of stock options in a privately-held company, Patents & Royalties: intellectual property rights; Rafael Pharmaceuticals: Other: grant support, Research Funding; Genentech: Consultancy, Honoraria, Other: grant support, Research Funding; Cellectis: Other: grant support; Calithera: Other: grant support, Research Funding; Agios: Other: grant support, Research Funding; Forty Seven: Other: grant support, Research Funding. Haferlach: MLL Munich Leukemia Laboratory: Other: Part ownership. Andreeff: Senti-Bio: Consultancy; ONO Pharmaceuticals: Research Funding; Glycomimetics: Consultancy; Aptose: Consultancy; Breast Cancer Research Foundation: Research Funding; Oxford Biomedica UK: Research Funding; Karyopharm: Research Funding; Medicxi: Consultancy; Amgen: Research Funding; AstraZeneca: Research Funding; Daiichi-Sankyo: Consultancy, Research Funding; Syndax: Consultancy; Novartis, Cancer UK; Leukemia & Lymphoma Society (LLS), German Research Council; NCI-RDCRN (Rare Disease Clin Network), CLL Foundation; Novartis: Membership on an entity's Board of Directors or advisory committees; Reata, Aptose, Eutropics, SentiBio; Chimerix, Oncolyze: Current holder of individual stocks in a privately-held company.